
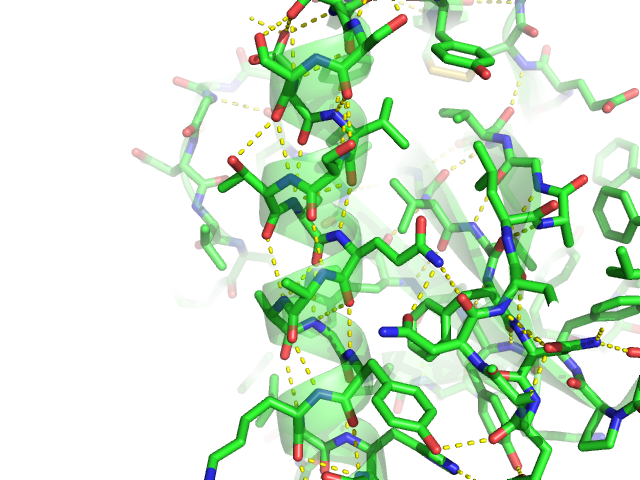
With such rich data at hand, we can gain deep insights into how ligands interact with their protein targets ( 2, 3). Binding of a ligand to its host protein requires a specific arrangement of attractive, typically non-covalent contacts between both molecules. The Protein Data Bank (PDB) ( 1) hosts nearly 100 000 deposited protein structures, with over 75% of them solved in complex with a small molecule ligand. PLIP's command-line mode allows for high-throughput interaction profiling.
#Pymol tutorial how to show ligand interactions full
The full python source code is available for download on the website. PLIP stands out by offering publication-ready images, PyMOL session files to generate custom images and parsable result files to facilitate successive data processing. It returns a list of detected interactions on single atom level, covering seven interaction types (hydrogen bonds, hydrophobic contacts, pi-stacking, pi-cation interactions, salt bridges, water bridges and halogen bonds). In contrast to other tools, the rule-based PLIP algorithm does not require any structure preparation. The input is either a Protein Data Bank structure, a protein or ligand name, or a custom protein–ligand complex (e.g.

Here, we present the protein–ligand interaction profiler (PLIP), a novel web service for fully automated detection and visualization of relevant non-covalent protein–ligand contacts in 3D structures, freely available at /plip-web. However, comprehensive tools are not freely available to the research community. The characterization of interactions in protein–ligand complexes is essential for research in structural bioinformatics, drug discovery and biology.


 0 kommentar(er)
0 kommentar(er)
